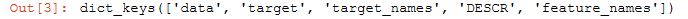Did you know that you can navigate the posts by swiping left and right?
Creating Artificial Neural Network With Python
24 Apr 2018
. category:
tech
.
Comments
#tutorial
Scikit Learn
In order to follow along with this tutorial, you’ll need to have the latest version of SciKit Learn installed! It is easily installable either through pip or conda, but you can reference the official installation documentation for complete details on this.
Data
We’ll use SciKit Learn’s built in Breast Cancer Data Set which has several features of tumors with a labeled class indicating whether the tumor was Malignant or Benign. We will try to create a neural network model that can take in these features and attempt to predict malignant or benign labels for tumors it has not seen before. Let’s go ahead and start by getting the data!
from sklearn.datasets import load_breast_cancer
cancer = load_breast_cancer()
}
This object is like a dictionary, it contains a description of the data and the features and targets:
cancer.keys()
# Print full description by running:
# print(cancer['DESCR'])
# 569 data points with 30 features
cancer['data'].shape
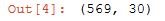 Let’s set up our Data and our Labels:
Let’s set up our Data and our Labels:
X = cancer['data']
y = cancer['target']
Train Test Split
Let’s split our data into training and testing sets, this is done easily with SciKit Learn’s train_test_split function from model_selection: Let’s set up our Data and our Labels:
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y)
Data Preprocessing
The neural network may have difficulty converging before the maximum number of iterations allowed if the data is not normalized. Multi-layer Perceptron is sensitive to feature scaling, so it is highly recommended to scale your data. Note that you must apply the same scaling to the test set for meaningful results. There are a lot of different methods for normalization of data, we will use the built-in StandardScaler for standardization.
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
# Fit only to the training data
scaler.fit(X_train)
# Now apply the transformations to the data:
X_train = scaler.transform(X_train)
X_test = scaler.transform(X_test)
Training the model
Now it is time to train our model. SciKit Learn makes this incredibly easy, by using estimator objects. In this case we will import our estimator (the Multi-Layer Perceptron Classifier model) from the neural_network library of SciKit-Learn!
from sklearn.neural_network import MLPClassifier
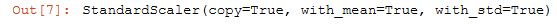
mlp = MLPClassifier(hidden_layer_sizes=(30,30,30))
Now that the model has been made we can fit the training data to our model, remember that this data has already been processed and scaled:
mlp.fit(X_train,y_train)

Predictions and Evaluation
Now that we have a model it is time to use it to get predictions! We can do this simply with the predict() method off of our fitted model:
predictions = mlp.predict(X_test)
Now we can use SciKit-Learn’s built in metrics such as a classification report and confusion matrix to evaluate how well our model performed:
from sklearn.metrics import classification_report,confusion_matrix
print(confusion_matrix(y_test,predictions))

print(classification_report(y_test,predictions))
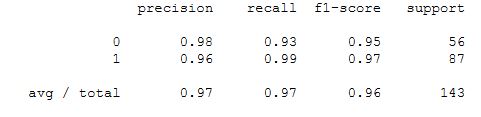 Looks like we only misclassified 3 tumors, leaving us with a 98% accuracy rate (as well as 98% precision and recall). This is pretty good considering how few lines of code we had to write! The downside however to using a Multi-Layer Preceptron model is how difficult it is to interpret the model itself. The weights and biases won’t be easily interpretable in relation to which features are important to the model itself.
Looks like we only misclassified 3 tumors, leaving us with a 98% accuracy rate (as well as 98% precision and recall). This is pretty good considering how few lines of code we had to write! The downside however to using a Multi-Layer Preceptron model is how difficult it is to interpret the model itself. The weights and biases won’t be easily interpretable in relation to which features are important to the model itself.